HLA Typing Service
Technical Information.
Introduction
A high resolution, HLA typing result applicable to most clinical, research and pharmacological requirements can be achieved without the need for sequencing.
The aim of this section is to provide some fundamental information on HLA nomenclature, allele frequencies and allele filters, and to explain how high resolution results are achieved using the MCD typing service.
HLA Nomenclature and Levels of Resolution
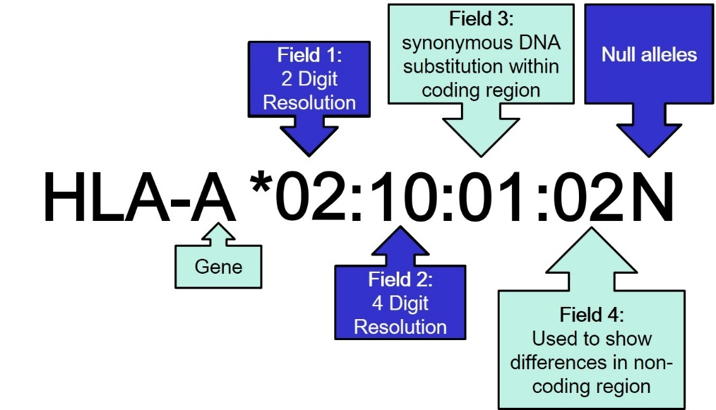
The WHO nomenclature (Figure 1) starts with the name of the gene locus followed by 4 fields indicating different levels of variation in the DNA sequence and the resulting protein. The fields are separated by a colon. The first field defines the allele group that usually corresponds to the serologically defined specificity of the HLA protein. Referring to the older nomenclature resolution on this level is called 2 digit (2D) resolution. The second field indicates differences in the DNA sequence that lead to a difference in the amino acid sequence of the resulting protein. The second field needs to be resolved to achieve 4 digit (4D) resolution. The third field is used to indicate synonymous DNA substitutions in the coding region and the fourth field refers to differences in the non-coding regions. Since differences in the third and fourth field do not have any influence on the resulting protein, it is usually not required to resolve them. At the end of the name for an HLA allele there may be a suffix indicating changes in the expression of the protein (N=not expressed Null allele, L= Low expression, Q= expression questionable). Null alleles are clinically relevant and usually need to be excluded or confirmed.
Figure 1: WHO nomenclature for HLA alleles
HISTO SPOT SSO Targets the Antigen Binding Domains
HLA class I proteins (HLA-A, -B & -C) are encoded by 8 exons and HLA class II proteins (HLA-DRB, -DQB & -DPB) are encoded by 6 exons. HISTO SPOT SSO targets the polymorphisms contained within those exons that encode for antigen binding domains of HLA molecules. For class I alleles this is exon 2 & 3 and for class II it is exon 2 only.
https://en.wikipedia.org/wiki/Human_leukocyte_antigen
HISTO SPOT SSO doesn’t distinguish HLA alleles having nucleotide sequences that encode the same protein sequence for the peptide binding domains. Alleles with identical antigen binding sites are designated together as P-Groups http://hla.alleles.org/alleles/p_groups.html, being unable to distinguish them doesn’t preclude the ability to generate a high resolution HLA type.
High Resolution Typing
The definition of high resolution in the European Federation of Immunogenetics (EFI) standards (V6.3) requires the identification of HLA alleles that encode the same protein sequence within the antigen binding site (P-groups). Specifically, the following requirements have to be met:
- First and second field of the WHO nomenclature resolved.
- All ambiguities resulting from polymorphisms in exon 2 and 3 for class I and from exon 2 for class II resolved.
- All ambiguities resolved that encompass a null allele wherever the polymorphism is located, unless the presence of an expressed antigen on the cell can be demonstrated (e.g. by serology).
Allele Frequencies and Filters
The Immuno Polymorphism Database: Release 3.26.01 (03 Nov 2016), has recognised over 15,600 HLA alleles. For full details see www.ebi.ac.uk/ipd/imgt/hla. However, most of these alleles have only been identified once or twice in the whole world. For this reason a group of international histocompatibility and immunogenetics researchers have defined and published the common and well-documented (CWD) alleles catalogue which identifies that subset of HLA alleles for which the frequencies are well known or which have been identified multiple times through the use of sequence-based typing methods http://igdawg.org/cwd.html. They have defined the following categories for frequent and rare alleles:
- Common (C): 415 alleles allele frequency > 0.001% in reference populations of at least 1500 individuals.
- Well-documented (WD): 707 alleles detected by SBT 5 times in unrelated individuals or detected by SBT 3 times and observed in a specific haplotype in unrelated individuals.
- Rare: All other alleles not defined as C or WD.
Our HISTO MATCH analysis software enables results to be analysed using a common (C) allele & a common and well documented (CWD) allele filter as defined above. A study was undertaken to show the resolution of the HISTO SPOT® kits. The study showed that when using the C filter >95% of samples typed were unambiguously resolved (only 2 common alleles found) enabling high resolution typing at the C allele level for P groups.
Reference:
1. Common and well-documented HLA alleles: 2012 update to the CWD catalogue; Mack S.J. et al. Tissue Antigens 2013, Vol 81, Issue 4, 183-257.
To discuss your purchase requirements in complete confidence, please Contact us using our secure enquiry form, and one of our management team will get back in touch.
Click any of the following links for further product information: